click here to download project abstract
click here to download project abstract
ABSTRACT
DEEP learning with convolution neural networks (CNNs) has achieved state-of-the-art performance for automated medical image segmentation. However, automatic segmentation methods have not demonstrated sufficiently accurate and robust results for clinical use due to the inherent challenges of medical images, such as poor image quality, different imaging and segmentation protocols and variations among patients. Interactive segmentation often requires image specific learning to deal with large context variations among different images but current CNNs are not adaptive to different test images, as parameters of the model are learned from training images and then fixed in the testing stage without image specific adaptation.
The Proposed system focus on interactive tumor segmentation of medical image sequences using deep neural network. The proposed work utilizes pattern based classification using neural network function. Adaptive Hierarchical motion segmentation is designed in the proposed area.
Abstract:
The project “Tumour Detection Using Interactive Image Segmentation and CNN” aims to enhance the accuracy and efficiency of tumor detection in medical images through the integration of Interactive Image Segmentation (IIS) and Convolutional Neural Networks (CNNs). The existing system for tumor detection often relies on traditional image processing techniques, which may lack precision in complex cases. The proposed system leverages state-of-the-art deep learning methods for improved tumor identification.
Existing System:
The current tumor detection systems employ conventional image processing algorithms, which may result in suboptimal accuracy, especially in cases of intricate tumor boundaries.
Proposed System:
The proposed system integrates Interactive Image Segmentation techniques with CNNs to improve accuracy and enable user interaction in the segmentation process. This synergy enhances the precision of tumor detection, allowing for more effective diagnosis and treatment planning.
System Requirements:
- CPU: Quad-core processor or higher
- RAM: 8GB or more
- GPU: NVIDIA GeForce GTX 1060 or equivalent
- Storage: 256GB SSD or higher
- Operating System: Windows 10 or Linux
Algorithms:
- Interactive Image Segmentation: GrabCut or GraphCut
- Deep Learning: Convolutional Neural Networks (CNNs)
Hardware and Software Requirements:
- Hardware: PC with recommended specifications
- Software: Python, TensorFlow, OpenCV, scikit-learn, Flask (for web UI)
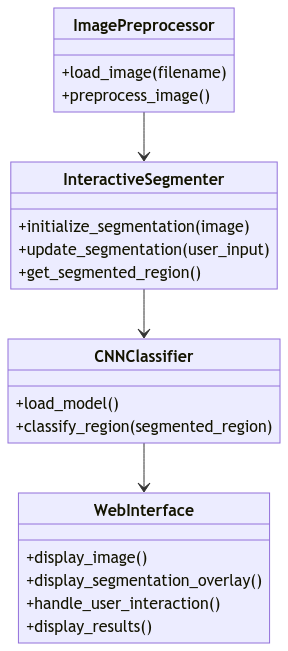
Architecture:
The system architecture comprises two main modules: Interactive Image Segmentation and CNN-based Tumor Detection. The Interactive Image Segmentation module allows user interaction for refining tumor boundaries, while the CNN module performs classification based on the segmented regions.
Technologies Used:
- Programming Language: Python
- Libraries: TensorFlow, OpenCV, scikit-learn
- Web UI: Flask for building a user-friendly interface
Web User Interface:
The system includes a web-based interface developed using Flask, allowing users to upload medical images, interact with the segmentation process, and view the detected tumor regions.
Graph LR Code for Architecture:

Class Diagram:
Sequence Diagram

use case diagram
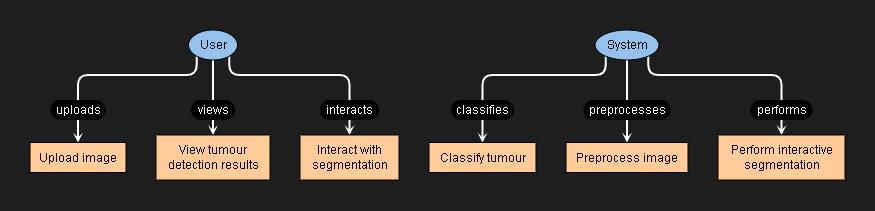
This project amalgamates cutting-edge techniques in interactive image segmentation and deep learning, providing a robust system for tumor detection in medical images. The combination of user interaction and CNNs enhances the accuracy and interpretability of the results, contributing to improved medical diagnostics.




